Appearance
Import Tutorial
WARNING
The import feature has been signifcantly overhauled. VS Code workspace storage will no longer be used and there will likely be further overhauls before reaching a final form.
Introduction
Importing references is more facile for documenting information flow in an notebook document. Previously, imports were centrally managed by the IBM materials Notebook extension which will look for the /lib workspace folder for JSON files. Now, with the latest update to CMDL, imports can directly reference any file within the current workspace. However, the /lib director is still recommended for holding files containing references to import.
The current version of CMDL now supports text files with the file extension .cmdl. These files types of files can be used both in the same way as the notebook files, albeit without execution/interpretation of the CMDL. These are also convient for definition of common chemical entities, polymer structures, continuous-flow reactors, or other items that may be shared between many notebooks or other text files. Prior versions of CMDL used JSON files to store shared data in the /lib folder. These have been removed in favor of using .cmdl files for imports.
Here we will demonstrate the use of the import keyword in the context of a chain extension experiment in polymer chemistry.
Setup
To demonstrate the import feature of IBM Materials Notebooks, make a copy of the batch tutorial notebook and save it as batch_exp_import.cmdnb in the same workspace used for the batch experiment tutorial. Or, you may copy the completed notebook provided in the /examples/batch_tutorial directory on the IBM Materials Notebook GitHub repository.
Finally, we will need ensure to provide the workspace with .cmdl files of chemicals and polymer graphs to import. These files can be copied from the /lib directory in the batch tutorial. Once these files have been copied over, close and reload the workspace folder.
Note: If you have completed and ran successfully the notebook from the batch tutorial then the needed file from the /lib directory should already be present.
Converting the Batch Experiment to use Imports
As a start, lets first convert the copied notebook to use imported references instead of explicitly defining each chemical, material, or graph reference.
With the batch_exp_import.cmdnb notebook open, delete cells containing all chemical, fragments, and polymer graph definitions for:
THFlLactidekOtBufragmentsPEG_BASEPEG_PLLA_Base
Now you should be seeing a lot of errors appear as the CDML compiler will recognize that many references are now missing. In a new cell at the top of the notebook, add the following imports:
cmdl
import tetrahydrofuran from "./lib/chemicals.cmdl";
import potassium_tert-butoxide from "./lib/chemicals.cmdl";
import dimethyl-1-4-dioxane-2-5-dione from "./lib/chemicals.cmdl";The CMDL syntax for imports utilizes the import and from keywords. Available imports are suggested through a VS Code completion provider.
Import Aliasing
Similar to variables in programming languages, reference names should be unique and not start with a number. All references globally scoped (along with other named groups such as char_data or reaction). However, many chemical names—especially IUPAC names—are poorly suited to be handles inside CDML or other languages, given the propensity to be overly long or contain many non-alphanumeric characters.
To get around this, we can use import aliasing using the as keyword to provide a more convient abbreviation for different reagent names. Aliases remain local to experiment notebook and the fullname, along with its alias are written to the experiment output.
Change the above example to use aliasing:
cmdl
import tetrahydrofuran as THF from "./lib/chemicals.cmdl";
import potassium_tert-butoxide as kOtBu from "./lib/chemicals.cmdl";
import dimethyl-1-4-dioxane-2-5-dione as lLactide from "./lib/chemicals.cmdl";All of these references are defined in the chemicals.cmdl file in the /lib directory of the workspace, which is automatically parsed and loaded upon activation of the extension in a new workspace. To complete the setup for the chain extension experiment in this tutorial, let's examine importing other references.
Experiment Setup
First let's define the polymer reference for our targeted product of the chain extension reaction. This gives a fresh instance to attach new results to based off of the outcome of the chain extension reaction. Rather than re-defining the entire polymer graph, let's import the polymer graph and add it to a new polymer reference. Add the following line to the cell with imports:
cmdl
import tetrahydrofuran as THF from "./lib/chemicals.cmdl";
import potassium_tert-butoxide as kOtBu from "./lib/chemicals.cmdl";
import dimethyl-1-4-dioxane-2-5-dione as lLactide from "./lib/chemicals.cmdl";
import PEG_PLLA from "./lib/polymerGraphs.cmdl";Next, lets define the polymer reference and attach the existing degree_poly property for the poly(ethylene glycol) repeat unit in the graph representation.
cmdl
import tetrahydrofuran as THF from "./lib/chemicals.cmdl";
import potassium_tert-butoxide as kOtBu from "./lib/chemicals.cmdl";
import dimethyl-1-4-dioxane-2-5-dione as lLactide from "./lib/chemicals.cmdl";
import PEG_PLLA from "./lib/polymerGraphs.cmdl";
polymer mPEG-PLLA {
structure: @PEG_PLLA;
@PEG_PLLA.PEG_Block.PEO {
degree_poly: 112.8;
};
}Importing the Product from the Batch Experiment
To complete the references we can import the product form the batch experiment. Here the import will be coming from one of the products from batch_experiment.cmdnb directory. Imports from this directory are labeled by their sample id given to them in the experiment notebook to help distinguish them from other product(s) from the same experiment and products from different experiments in the same workspace.
We can modify the imports to include mPEG-PLLA-Test-I-123A which would correspond to the intermediate product from the prior batch experiment.
cmdl
import tetrahydrofuran as THF from "./lib/chemicals.cmdl";
import potassium_tert-butoxide as kOtBu from "./lib/chemicals.cmdl";
import dimethyl-1-4-dioxane-2-5-dione as lLactide from "./lib/chemicals.cmdl";
import PEG_PLLA from "./lib/polymerGraphs.cmdl";
import mPEG-PLLA-Test-I-123A from "./batch_experiment.cmdnb";
polymer mPEG-PLLA {
structure: @PEG_PLLA_Graph;
@PEG_PLLA_Graph.PEG_Block.p_PEO {
degree_poly: 112.8
};
}Note that we do not assign a value for the DPn of the lactide block in the mPEG-PLLA definition even though technically it is already somewhat defined by the starting material mPEG-PLLA-Test-I-123A. Since this block is being extended with an identical monomer, we can add the DPn during the characterization phase for this case. Were we creating a block copolymer with a different monomer we would then probably define the DPn.
cmdl
import tetrahydrofuran as THF from "./lib/chemicals.cmdl";
import potassium_tert-butoxide as kOtBu from "./lib/chemicals.cmdl";
import dimethyl-1-4-dioxane-2-5-dione as lLactide from "./lib/chemicals.cmdl";
import PEG_PLLA from "./lib/polymerGraphs.cmdl";
import mPEG-PLLA-Test-I-123A from "./batch_experiment.cmdnb";
polymer mPEG-PLLA {
structure: @PEG_PLLA_Graph;
@PEG_PLLA_Graph.PEG_Block.p_PEO {
degree_poly: 112.8
};
}Executing the the cell will complete the import process.
Defining the Chain Extension Reaction
With the references properly defined. We can create the chain extension reaction group:
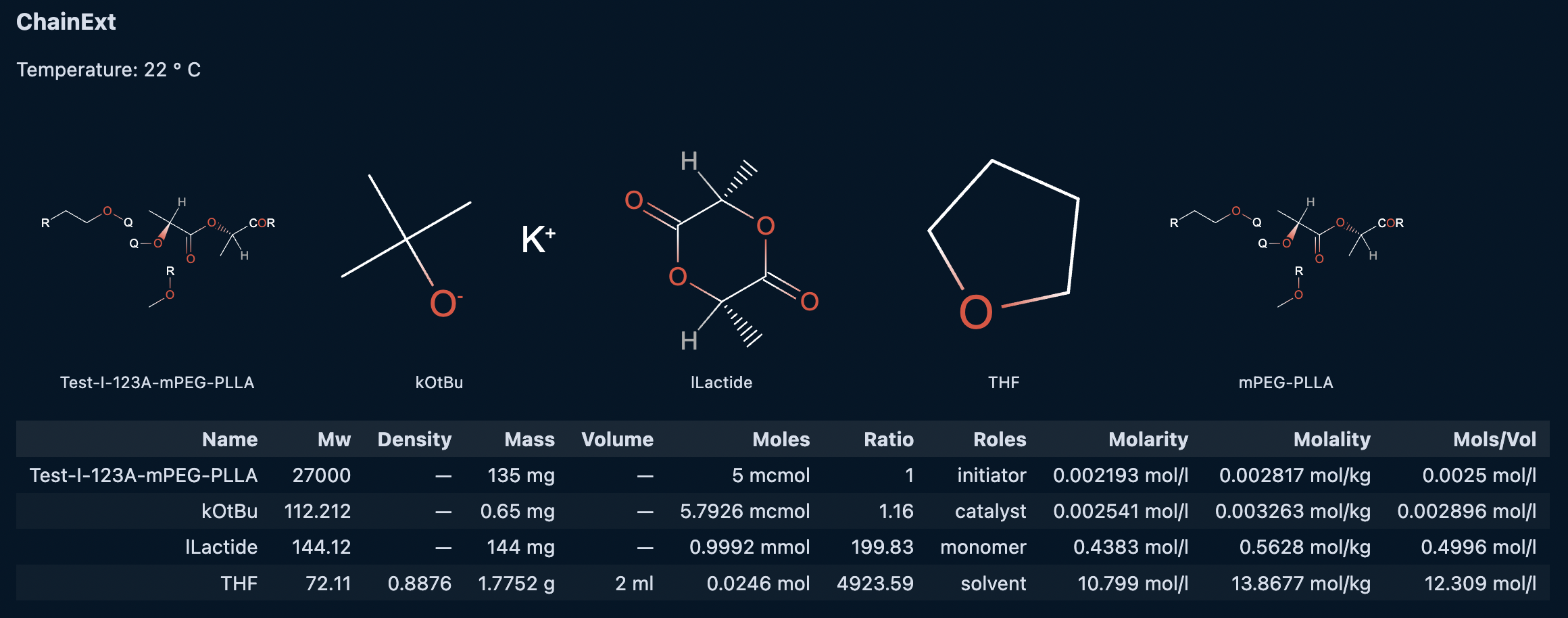
cmdl
reaction ChainExt {
temperature: 22 degC;
reaction_time: 10 s;
@mPEG-PLLA-Test-I-123A {
mass: 135 mg;
roles: ["initiator"];
};
@kOtBu {
mass: 0.65 mg;
roles: ["catalyst"];
};
@lLactide {
mass: 144 mg;
roles: ["monomer"];
};
@THF {
volume: 2 ml;
roles: ["solvent"];
};
@mPEG-PLLA {
roles: ["product"];
};
}Running the cell will produce an output similar to the following:
Defining the Characterization Data
With the reaction and protocol groups object in place, we can add a cell to define the results and also add the metadata of any characterization techniques. In a new cell add the following:
cmdl
char_data Test-II-123A-nmr {
sample_id: "test-123-A";
technique: "nmr";
time_point: 5 s;
@lLactide {
conversion: 80%;
};
@mPEG-PLLA.PEG_PLLA.Lactide_Block.Llac {
degree_poly: 347.5;
};
}
char_data Test-II-123A-gpc {
sample_id: "test-123-A";
technique: "gpc";
time_point: 5 s;
@mPEG-PLLA {
dispersity: 1.4;
mn_avg: 52000 g/mol;
};
}In the latest updates to CMDL, we can now start to import and reference characterization data files. Currently, only simple characterization data in .csv format are supported. Future versions will expand support to other types of characterization data. In the batch tutorial folder, there is a sample GPC trace in a /data folder, this folder can be copied into your tutorial workspace and the data can be imported by updating the import cell as follows:
cmdl
import tetrahydrofuran as THF from "./lib/chemicals.cmdl";
import potassium_tert-butoxide as kOtBu from "./lib/chemicals.cmdl";
import dimethyl-1-4-dioxane-2-5-dione as lLactide from "./lib/chemicals.cmdl";
import PEG_PLLA from "./lib/polymerGraphs.cmdl";
import mPEG-PLLA-Test-I-123A from "./batch_experiment.cmdnb";
import * as Test-123-gpc from "./data/test-123-gpc.csv";
polymer mPEG-PLLA {
structure: @PEG_PLLA_Graph;
@PEG_PLLA_Graph.PEG_Block.p_PEO {
degree_poly: 112.8
};
}Here we use the * syntax to simply indicate that full file is being imported and assigned an alias Test-123-gpc. Next we can specify which char_data sample the characterization data was generated.
cmdl
char_data Test-II-123A-nmr {
sample_id: "test-123-A";
technique: "nmr";
time_point: 5 s;
@lLactide {
conversion: 80%;
};
@mPEG-PLLA.PEG_PLLA.Lactide_Block.Llac {
degree_poly: 347.5;
};
}
char_data Test-II-123A-gpc {
sample_id: "test-123-A";
technique: "gpc";
time_point: 5 s;
file: @Test-123-gpc;
@mPEG-PLLA {
dispersity: 1.4;
mn_avg: 52000 g/mol;
};
}Running the cell will provide a preview of the data as follows:
WARNING
Importing and viewing characterization data is still under active development. Currently there is only support for .csv files for GPC traces. This will change in the very near future.
Next Steps
Now that we have completed batch experiments, we can move on to the final tutorial on how to define and execute continuous-flow experiments.